
A blog on data management and DataLad



This may be a bit of an unusual post for this blog, but it still fits into the bigger picture. So here we go… For years I wanted to have a digital photo frame. One of these devices that hang on the wall and make the thousands and thousand of pictures taken over many years visible. Although I can look at them on my phone, this would something that I need to do actively. A framed picture is just there, waiting for me. A picture on the wall is also an invitation to talk about it, much more than the relative privacy of a phone screen. ...

Prelude: Scientist in a data labyrinth As experimental neuroscientist in training, I often find myself caught between two worlds: the messy, exploratory world of data analysis where I try to make sense of the experimental data, often relying on a trial-and-error strategy, and the aspired structured, reproducible world of scientific publication where hopefully every figure will be exactly reproducible. Between these worlds lies a labyrinth of processing pipelines, half-written scripts, and the ever-present risk of breaking the working analysis while trying to improve it. The wandering in the labyrinth is rarely straight-forward – one is expected to hit many dead ends and discover other interesting distractions before finding the actual treasure – the key results that hopefully lead to a scientific publication or other forms of consolidated knowledge piece. I try to illustrate this metaphoric labyrinth that is my non-metaphoric reality in a diagram: ...

Git-annex continues to evolve. In this post, I want to look at two changes, one big and one small, introduced within the last year. Together, they make publishing files through Nextcloud much nicer. Specifically, it is now possible for a read-only shared Nextcloud folder to be a one-stop shop for cloning the dataset and getting file contents. This can be a useful setup for sharing (research) data: having the shared folder be a single point of access is convenient, and restricting write access is necessary to prevent unauthorized changes. ...

For the past 18 years I have been a GitHub user. It has been an extremely convenient platform for collaborating with many people from all over the world. What makes GitHub, and other platforms like it, particularly attractive is that they are typically way more accessible than any institutionally provided infrastructure (even if not without issues of its own). GitHub also provided an extremely reliable and stable infrastructure that encouraged and rewarded building on it. Again, from my personal experience, much more reliable and stable than lots of institutional infrastructures. ...
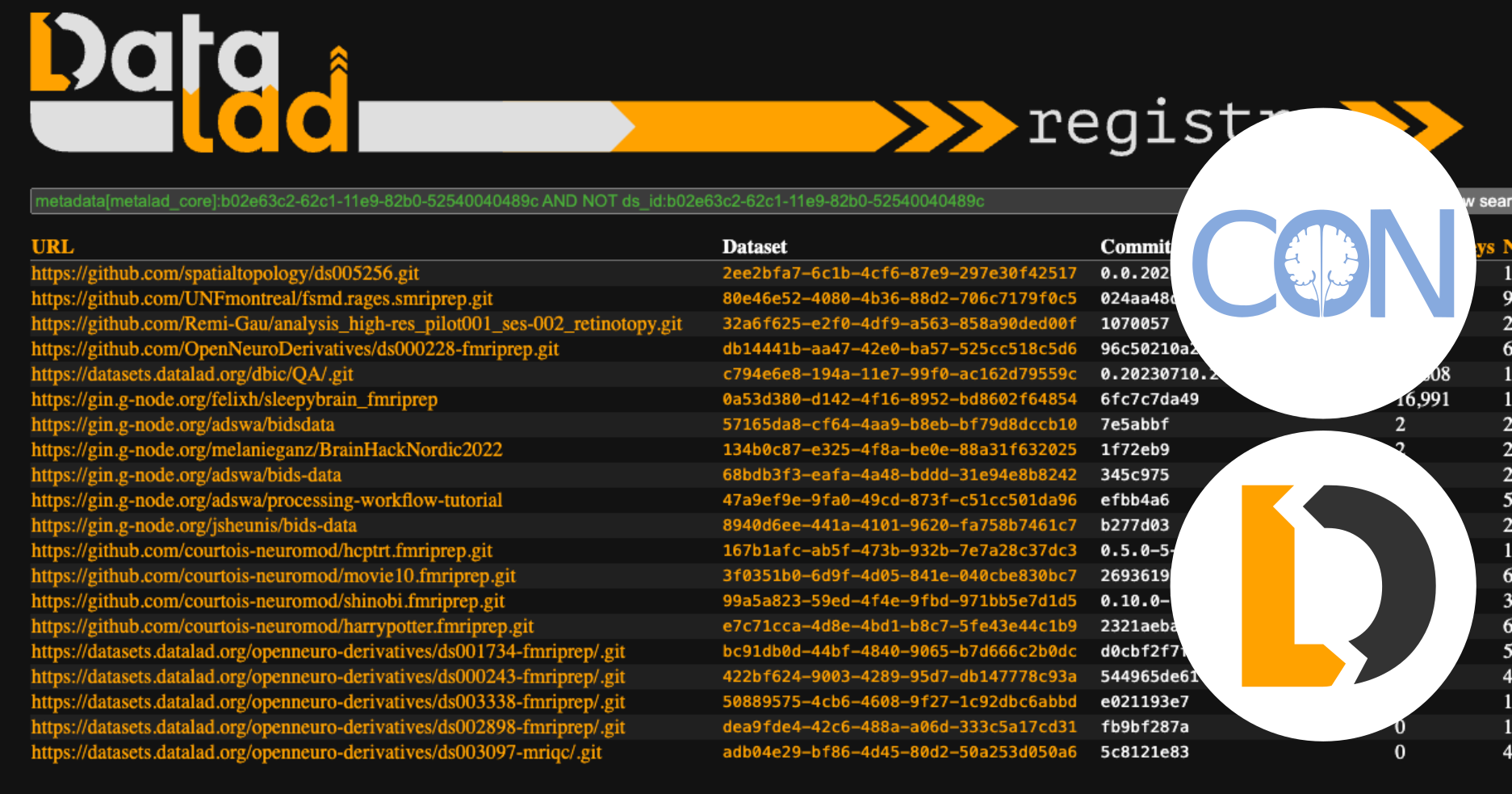
DataLad provides a platform for managing and uniformly accessing data resources. It also captures basic provenance information about data results within Git repository commits. However, discovering DataLad datasets or Git repositories that DataLad has operated on can be challenging. They can be shared anywhere online, including popular Git hosting platforms, such as GitHub, generic file hosting platforms such as OSF, neuroscience platforms, such as GIN, or they can even be available only within the internal network of an organization or just one particular server. We built DataLad-Registry to address some of the problems in locating datasets and finding useful information about them. (For convenience, we will use the term “dataset”, for the rest of this blog, to refer to a DataLad dataset or a Git repo that has been “touched” by the datalad run command, i.e. one that has “DATALAD RUNCMD” in a commit message.) ...