Prelude: Scientist in a data labyrinth
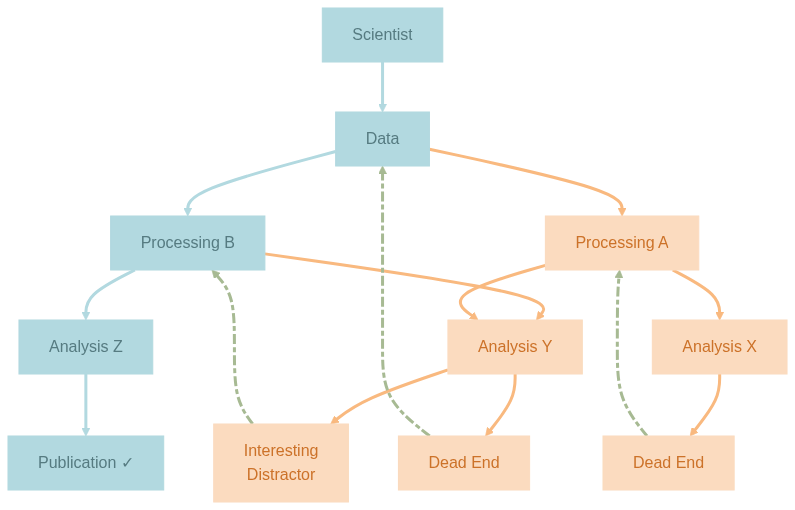
As experimental neuroscientist in training, I often find myself caught between two worlds: the messy, exploratory world of data analysis where I try to make sense of the experimental data, often relying on a trial-and-error strategy, and the aspired structured, reproducible world of scientific publication where hopefully every figure will be exactly reproducible. Between these worlds lies a labyrinth of processing pipelines, half-written scripts, and the ever-present risk of breaking the working analysis while trying to improve it. The wandering in the labyrinth is rarely straight-forward – one is expected to hit many dead ends and discover other interesting distractions before finding the actual treasure – the key results that hopefully lead to a scientific publication or other forms of consolidated knowledge piece. I try to illustrate this metaphoric labyrinth that is my non-metaphoric reality in a diagram:
So how do I navigate this mess? Ideally, I can (1) keep track of all my all tiny and big successes and failures, (2) do so without blowing up the project with unnecessary redundancy, (3) reuse or combine elements of (pre-)processing and analysis in different ways, (4) and (re-)visit and abandon or recover any state at any given time. In short, I want to experiment freely and safely in the space that is spanned by the data and the available (meaningful) manipulations without compromising reproducibility or creating unnecessary overheads.
Introduction: Better modular than regular
A useful concept that lights my way (though how enlighted I am not sure am I) is YODA’s Organigram on Data Analysis that captures the most key principle of managing reproducible complex scientific workflow involving digital objects: modularity.
The first level of modularity is fairly straight-forward: each component of the workflow should be a stand-alone module on its own (-> reusable, combinable), e.g. raw data, derived data, code, containers. In the diagram above each node is a module as such. Yes, the scientist is also a stand-alone module (so I hope :D). To achieve this level of modularity, it is enough to create a repository/dataset for each component using a version control system such as Git (only for code or other small files) or DataLad. Datalad is built on Git and Git-Annex and can handle both code and data (or basically anything you care about).
The second level of modularity is less obvious, but the actually key for achieving reproducible workflows and not just a static collection of version-controlled data: how to move from one state to another or in other words, how all but raw data come into existence (-> provenance-tracking) including that figure you presented in the last lab meeting back in January and can’t find the code to reproduce it (ha, gotcha!). In the diagram this is represented by the directed edges with the respective parent and child. Take the connection Data -> Processing A, a module in YODA sense is the mapping from raw data (inputs) to processed data (outputs) with the processing pipeline A (code). In practice, we now link two or more submodules/subdatasets together in a functional module aka superdataset (-> hierarchical). But not just that, we can now version-control the manipulations on data, as well, using the datalad (containers-) run command that ensures full provenance tracking. At this level of modularity, we likely quickly run into dataset-nesting since any functional module can also be the input to another functional module downstream as illustrated in the diagram, e.g. processed data after manipulation A is provided to Analysis X and Analysis Y as inputs. Per modularity principle, this requires the installation of Processing A as a subdataset inside Analysis X/Y. A person working only on Analysis X/Y will see Processing A only once, but a person working on both Processing A and Analysis X/Y will see Processing A twice – because once per functional module! How to avoid unnecessary overheads here? Keep Processing A as the canonical instance in the functional module Data -> Processing A, and install an ephemeral instance of it as subdataset in Processing A -> Analysis X/Y. This way, both instances share the same annex content via symlinks (-> no redundancy). Pretty smart, right?
Is there a third level of modularity? I would argue yes! If the first level is components (raw data, code, etc.) and the second is provenance (tracking how components transform into one another), then the third level is orchestration - how we safely and efficiently execute these transformations, especially when they need to happen in parallel, in isolation, or across different environments. In our labyrinth metaphor, we now have:
- Nodes = Components (data, code, containers)
- Nodes + Edges = Provenance (how nodes transform)
- Paths = Execution strategies (how we navigate the graph)
This third level addresses questions like:
- How do I run 100 analyses in parallel? -> see this heavyweight but beautiful fairly-big-processing-workflow
- How do I ensure the same analysis runs identically on my laptop, the cluster, and in 6 months where some packages might have updated? -> see DataLad Handbook: 7.2. Computational reproducibility with software containers
- How do I explore multiple states simultaneously without them interfering with each other, and at no cost, so that the expedition can be afforded daily?
This blog post is all about the third question. Before we dig in, have you noticed that something is missing from my wishlist? Let’s briefly return to the metaphoric labyrinth that in reality is a decision tree: We’ve got multiple variants on each level, i.e. two ways of (pre-)processing, three ways of analyzing, and four potential outcomes. In the very end, after a long, exhaustive, and hopefully successful wandering we only need to keep and reproduce the path Data -> Processing B -> Analysis Z -> Publication (colored in Morning Glacier). Do we really want to keep all the less fruitful attempts (colored in Rosy Horizon), especially if they are actually derived from each other, and not completely new stuff? Very good question! After hitting the first Dead End, we might not backtrack (dashed lines colored in Sagebrush Drift) to Processing A, but to Analysis X instead, and derive Analysis Y from it. We might also derive Analysis Z from Analysis Y, and Processing B from Processing A. What do we end up with? Classical versioning of the same thing! So instead of investing stand-alone modules in these variants and blowing up our project with redundancy, we can simply leave them in the repo history and let Git manage them (-> time-traveling). For a linear history we can use git tags to label the specific states/commits for later retrieval. If we decide to continue developing from there in a divergent fashion at a later time, we can create a specific branch and develop there. For our diagram above, all components at the same level can indeed be combined into one module, with the node colored in Morning Glacier being the present (or in my case the future since I haven’t found the treasure yet) or the final state, and all nodes colored in Rosy Horizon being the past (or in my case the present since I’m still in the labyrinth) or the (git) history. Now, that we have enabled time-traveling in our project, which means I’m free to checkout any state at any time, we can tackle the real challenge: how do we visit multiple states simultaneously (-> parallel universes aka Everything Everywhere All At Once)?
Origin Story: When YODA’s wisdom grows on trees
It all started at the recent Distribits meeting, when Yarik introduced the Git worktrees to me upon my question: “How can I continue working on my dataset when I’m running a processing script that takes a long while?”. To ensure reproducibility, datalad run command requires the datasets to be clean. Executing a series of datalad runcommands effectively blocks me from working on any of the involved components. A git worktree is a built-in Git feature that allows the creation of multiple, separate working directories from a single Git repository. Each worktree is linked to a specific branch or commit, meaning one can check out and work on different branches simultaneously without affecting the others. The cherry on the top: it provides isolation without duplication. All worktrees share the same core repository history and data. It sounds all fabulous and magical, so I set out to achieve a workflow using git worktrees to have the best of both worlds: fast iteration during development and clean reproducibility for batch processing.
| Context | State | Purpose | Data Access |
|---|---|---|---|
| Main Worktree | Active development | Trying new analyses, debugging, exploration | Shared annex symlinks |
runs Worktree | Frozen, stable state | Batch processing, reproducible execution | Shared annex symlinks |
Now that I’ve introduced the most important concepts and the objective, I’ll leave you alone with the report of the journey that I jotted down along the process with almost no post-hoc modification. As original as it can be ;) Enjoy!
For the impatient: Here is where the fun starts
Meng_Psychedelics/ (superdataset)
├── raw/
│ ├── L5b_2p/ (subdataset)
│ └── L5b_bpod/ (subdataset)
├── derived/
│ └── L5b/ (subdataset)
│ ├── inputs/
│ │ ├── L5b_2p/ (subdataset → ../../raw/L5b_2p)
│ │ └── L5b_bpod/ (subdataset → ../../raw/L5b_bpod)
│ ├── code/ (subdataset)
│ ├── containers/ (subdataset)
│ ├── 01_suite2p/
│ ├── ...
│ └── 04_dataframes/
└── ...
The active work in my project happens in the derived/L5b subdataset that consumes raw data as inputs and produces multiple intermediate outputs, e.g. 01_suite2p … 04_dataframes. The subdataset code is a pure git repo and I use Jujutsu for active development. I encountered jj the same time as I heard of the worktrees (check out Lorenz’s talk on Radicle and his experience with Jujutsu) and fell in love instantly (read more here). After a rather nerve-wracking experience of mixing datalad with jj - it was like Schrödinger’s cat, everything was simultaneously staged and unstaged 😱 - I restrict jj usage to code, and continue using datalad for managing all subdatasets with annexed content, orchestrating across nested datasets as a whole, and for capturing provenance of all derived data and figures with the datalad (containers-) run command.
Neither ready to go big nor go home: worktrees instead of clones
Usually, I craft one datalad run command, test it, and then write a bash script to run it for multiple subjects and experiments. Here is when it becomes painful: once it’s running you can’t carry on working, because any modification will interrupt the next datalad run command. Ideally, I can run things in an isolated environment that is clean and stays untouched until the job finishes. Git worktrees come in handy, because they essentially access the same annexed content (so no time wasted in getting content for another clone), but live in a different directory on the disk. Worktrees share the annex and repo history, but not the state the repo is in. This allows me to checkout different branches simultaneously since they live on different worktrees.
Since at the moment there is no DataLad helper command to create a collection of nested worktrees (described in this issue) for each subdataset, I directly use git worktree add command to create a worktree for each subdataset inside derived/L5b:
# add a worktree on a new branch with the name `runs` for the superds
M/d/L5b (master)> git worktree add /mnt/Data/L5b-runs -b runs
Preparing worktree (new branch `runs`)
HEAD is now at fde26554f [DATALAD RUNCMD] plot rois, traces and F displots for mouse J, Lisuride
# repeat for each subdataset
M/d/L5b (master)> cd inputs/L5b_bpod
M/d/L/i/L5b_bpod (master)> git worktree add /mnt/Data/L5b-runs/inputs/L5b_bpod -b runs
Preparing worktree (new branch `runs`)
HEAD is now at e32a9bbf2c [DATALAD RUNCMD] Rename bpod files of mouse J, LSD with the missing date.
M/d/L/i/L5b_bpod (master)> cd ../L5b_2p
M/d/L/i/L5b_2p (master)> git worktree add /mnt/Data/L5b-runs/inputs/L5b_2p -b runs
Preparing worktree (new branch `runs`)
HEAD is now at 3bd0b284b2 rename interrupted recording
M/d/L/i/L5b_2p (master)> cd ../../code
M/d/L/code> git worktree add /mnt/Data/L5b-runs/code runs # here I skip -b because the branch already exists
Preparing worktree (checking out `runs`)
HEAD is now at ef5dafb generate_plots: plot rois, traces, and F displots
The branch runs in code/ has already been created, when I got ready with the code:
# the new datalad run command is ready
M/d/L/code> git switch -c runs
Switched to a new branch `runs`
# `runs` points to the commit ef5dafb4: generate_plots
M/d/L/code> jj log
Done importing changes from the underlying Git repo.
@ turmruqs jiameng.wu@gmail.com 2025-12-03 13:38:37 f8743c11
│ (empty) (no description set) # <- the clean state we need
│ ○ tnxopmxz jiameng.wu@gmail.com 2025-12-03 13:38:37 2cd4912a
├─╯ bar plot of mice IJK # <- this is just another dev branch
○ vlwmszww jiameng.wu@gmail.com 2025-12-03 13:21:37 master?? runs git_head() ef5dafb4
│ generate_plots: plot rois, traces, and F displots # <- target commit
For all perspective jj users, having the empty change on top of the target commit is a necessary condition for a clean working tree and thus for datalad run to happen.
So the plan is to park the worktree runs at this clean state on top of the commit ef5dafb4, run the script generate_plots.py on multiple subjects and experiments, while continuing developing on dirty branches. Let’s see if that works out!
In the newly created worktree runs, I verify that it is in the right state that records the successful datalad run command:
# in worktree `runs`
/m/D/L5b-runs (runs)> git log --oneline
fde26554f (HEAD -> runs, master) [DATALAD RUNCMD] plot rois, traces and F displots for mouse J, Lisuride
Nice, I will first attempt a rerun! Oh wait, not so fast, I need to set up my Python virtual environment using a wonderful and fast uv tool. So I go:
cd code
uv sync
source .venv/bin/activate.fish
cd ..
Now, RERUN!
/m/D/L5b-runs (runs)> datalad rerun
[INFO ] run commit fde2655; (plot rois, traces...)
[INFO ] Making sure inputs are available (this may take some time)
unlock(ok): 05_visualization/240227J_Sim-Cre/displots/cell_01.png (file)
unlock(ok): 05_visualization/240227J_Sim-Cre/displots/cell_02.png (file)
unlock(ok): 05_visualization/240227J_Sim-Cre/displots/cell_03.png (file)
unlock(ok): 05_visualization/240227J_Sim-Cre/displots/cell_04.png (file)
unlock(ok): 05_visualization/240227J_Sim-Cre/displots/cell_06.png (file)
unlock(ok): 05_visualization/240227J_Sim-Cre/displots/cell_07.png (file)
unlock(ok): 05_visualization/240227J_Sim-Cre/displots/cell_08.png (file)
unlock(ok): 05_visualization/240227J_Sim-Cre/displots/cell_somata.png (file)
unlock(ok): 05_visualization/240227J_Sim-Cre/displots/cell_tuft.png (file)
unlock(ok): 05_visualization/240227J_Sim-Cre/rois/all_cells.png (file)
[1 similar message has been suppressed; disable with datalad.ui.suppress-simil[INFO ] == Command start (output follows) =====
Stacking the dataframe into trial format ...
Finished in 0.1674485206604004 second.
Plotting ROIs on top of the mean image for each cell ...
Plotting fluorescence traces for each cell across multiple ROIs ...
Plotting the distribution of the fluorescence values for each cell ...
[INFO ] == Command exit (modification check follows) =====
[18 similar messages have been suppressed; disable with datalad.ui.suppress-similar-results=off]
run(ok): /mnt/Data/L5b-runs (dataset) [python code/src/process2p/generate_plots...]
add(ok): 05_visualization/240227J_Sim-Cre/displots/cell_01.png (file)
add(ok): 05_visualization/240227J_Sim-Cre/displots/cell_02.png (file)
add(ok): 05_visualization/240227J_Sim-Cre/displots/cell_03.png (file)
add(ok): 05_visualization/240227J_Sim-Cre/displots/cell_04.png (file)
add(ok): 05_visualization/240227J_Sim-Cre/displots/cell_06.png (file)
add(ok): 05_visualization/240227J_Sim-Cre/displots/cell_07.png (file)
add(ok): 05_visualization/240227J_Sim-Cre/displots/cell_08.png (file)
add(ok): 05_visualization/240227J_Sim-Cre/displots/cell_somata.png (file)
add(ok): 05_visualization/240227J_Sim-Cre/displots/cell_tuft.png (file)
add(ok): 05_visualization/240227J_Sim-Cre/rois/all_cells.png (file)
[1 similar message has been suppressed; disable with datalad.ui.suppress-simil [18 similar messages have been suppressed; disable with datalad.ui.suppress-similar-results=off]
action summary:
add (ok: 28)
get (notneeded: 3)
run (ok: 1)
save (notneeded: 4)
unlock (ok: 28)
/m/D/L5b-runs (runs)> git log
fde26554f (HEAD -> runs, master) [DATALAD RUNCMD] plot rois, traces and F displots for mouse J, Lisuride
It completes without creating a new commit - the outputs are byte-for-byte identical to the previous run. This is where the magic happens: I’m running from a completely different directory, yet I don’t need to datalad get anything or datalad save results. The annex’s clever symlinks make the same data available in both worktrees, eliminating redundancy entirely! 🎩✨
Now it’s time to write that bash script, and to test how going forth and back between worktrees works. To not mix things up, I will never develop in the worktree runs, but only update it with what ever has been developed and become available on ‘master’ - well, actually we don’t care about branches in jj’s universe but anyway you get the gist.
#!/usr/bin/env bash
# This is the batch-processing script
set -u
# -------------------------------
# Target script
# -------------------------------
script="generate_plots.py"
# -------------------------------
# Inputs
# -------------------------------
subjects=("240226I" "240227J" "240229K")
drugs=("Ketamine" "LSD" "Lisuride" "Saline" "Saline2")
# -------------------------------
# Datalad RUNCMD
# -------------------------------
run_one() {
local subid="$1" drug="$2"
echo "=== ${subid} / ${drug} ==="
datalad run \
-m "Plot rois, traces and distribution of F_corrected for mouse ${subid}, ${drug}." \
-i "04_dataframes/${subid}_Sim-Cre/${drug}/plane0/roiset_suite2p/F.pkl" \
-i "01_suite2p/${subid}_Sim-Cre/${drug}/plane0/ops.npy" \
-i "01_suite2p/${subid}_Sim-Cre/${drug}/plane0/stat.npy" \
-o "05_visualization/${subid}_Sim-Cre/${drug}" \
"python code/src/process2p/${script} {inputs} {outputs}" || {
echo "ERROR: datalad run failed for ${subid} / ${drug}, but continuing..."
datalad save -m "run ${subid} / ${drug} failed, save and continue with the next one"
return 0 # Return success to continue the loop
}
}
# -------------------------------
# Main loop
# -------------------------------
failed_jobs=()
for subid in "${subjects[@]}"; do
for drug in "${drugs[@]}"; do
if ! run_one "${subid}" "${drug}"; then
failed_jobs+=("${subid}/${drug}")
echo "Recorded failure: ${subid} ${drug}"
fi
done
done
echo "All done."
I have added this bash script under pipelines/run_generate_plots.sh path and updated the README.md, but the git branch runs is still lagging behind and I’m in a HEAD detached state:
# back in the main worktree
/M/d/L/code> jj log
@ mxvrsvoq jiameng.wu@gmail.com 2025-12-04 19:57:42 708ab665
│ (empty) (no description set)
○ yknyuprl jiameng.wu@gmail.com 2025-12-04 19:56:49 git_head() 84312427
│ update README.md # <- another new commit
○ turmruqs jiameng.wu@gmail.com 2025-12-04 19:56:29 08065905
│ add bash script to run generate_plots.py # <- the new commit
│ ○ tnxopmxz jiameng.wu@gmail.com 2025-12-03 13:38:37 2cd4912a
├─╯ bar plot of mice IJK
○ vlwmszww jiameng.wu@gmail.com 2025-12-03 13:21:37 master?? runs ef5dafb4
│ generate_plots: plot rois, traces, and F displots # <- `runs` is still here
/M/d/L/code> git status
HEAD detached from refs/heads/runs
nothing to commit, working tree clean
Let’s update that branch and sew the HEAD back to runs. There are multiple ways to do that, including plain git commands, but since I’m exploring Jujutsu’s universe at the moment, I go:
/M/d/L/code [2]> jj bookmark set runs -r 'git_head()'
Moved 1 bookmarks to yknyuprl 84312427 runs* | update README.md
# now both `runs` and 'git_head()' point to the right commit
/M/d/L/code> jj log
@ mxvrsvoq jiameng.wu@gmail.com 2025-12-04 19:57:42 708ab665
│ (empty) (no description set)
○ yknyuprl jiameng.wu@gmail.com 2025-12-04 19:56:49 runs git_head() 84312427
│ update README.md # <- the right commit
○ turmruqs jiameng.wu@gmail.com 2025-12-04 19:56:29 08065905
│ add bash script to run generate_plots.py
│ ○ tnxopmxz jiameng.wu@gmail.com 2025-12-03 13:38:37 2cd4912a
├─╯ bar plot of mice IJK
○ vlwmszww jiameng.wu@gmail.com 2025-12-03 13:21:37 master?? ef5dafb4
│ generate_plots: plot rois, traces, and F displots
Back in worktree runs, I confirm the branch has been updated for code and the superdataset also registers the update:
# in worktree `runs`
/m/D/L/code (runs)> git log --oneline
8431242 (HEAD -> runs, refs/jj/keep/843124271281fc8e8a7a3ceff202a4fa6f0a08a8) update README.md
0806590 (refs/jj/keep/0806590504da65021ec465ddfac2fb3c7bc2c977) add bash script to run generate_plots.py
ef5dafb (refs/jj/keep/ef5dafb4ac4b7e9e2c6854cff726bede0c47ac53, master) generate_plots: plot rois, traces, and F displots
/m/D/L/code (runs)> cd ..
# in the superds of the same worktree
m/D/L5b-runs (runs)> datalad status
modified: code (dataset) # need to save this modification
Should I also save this update in my main worktree? No, I will only save it in this worktree and then update the main worktree when the plots are ready! (What’s datalad’s behavior with branches anyway?)
Seasoning with a pinch of Jujutsu magic
I’m almost ready to go, except that I have to amend another modification to the bash script. Out of curiosity I decide to use jj squash to add this modification to the previous change instead of creating a new commit. I observe that the commit ID changes, and that runs still points to the correct commit:
# in main worktree
/M/d/L/code> jj squash --into turmruqs
Rebased 1 descendant commits
Working copy (@) now at: oqwzryko 8af9e583 (empty) (no description set)
Parent commit (@-) : yknyuprl 25eff5ce runs | update README.md
# the commit ID associated with the change ID 'turmruqs' has changed from '08065905' to '42c0112b'
/M/d/L/code> jj log
@ oqwzryko jiameng.wu@gmail.com 2025-12-04 20:18:57 8af9e583
│ (empty) (no description set)
○ yknyuprl jiameng.wu@gmail.com 2025-12-04 20:18:57 runs git_head() 25eff5ce
│ update README.md # <- the child commit changed automatically
○ turmruqs jiameng.wu@gmail.com 2025-12-04 20:18:57 42c0112b
│ add bash script to run generate_plots.py # <- modification squashed into this commit
How does this change of history affect the other worktree?
# in worktree `runs`
/m/D/L/code (runs)> git log --oneline
25eff5c (HEAD -> runs, refs/jj/keep/25eff5cef05040cd0d54e4cfb36107c8f7708d0b) update README.md
42c0112 (refs/jj/keep/42c0112b40b45d58ce5d86bb57061ab5d0387d82) add bash script to run generate_plots.py
ef5dafb (refs/jj/keep/ef5dafb4ac4b7e9e2c6854cff726bede0c47ac53, master) generate_plots: plot rois, traces, and F displots
That’s fantastic! I sneaked in a new modification without running any datalad save command and get the clean worktree for free! Hurrraaayy!!! But wait, git and datalad does detect the change:
# still in worktree `runs`
/m/D/L/code (runs)> datalad status
modified: pipelines/run_generate_plots.sh (file) # <- that's the batch-processing script
/m/D/L/code (runs)> git status
On branch runs
Changes to be committed:
(use "git restore --staged <file>..." to unstage)
modified: pipelines/run_generate_plots.sh
It is because the file has in fact changed due to the jj squash operation. I changed the history and there is a mismatch with the presence. All I need to do is to reset it to runs with git reset --hard runs.
The trade-off between efficiency and reproducibility: You win some you loose some
The script fails, and I try to rerun this one command that worked earlier, it fails as well:
/m/D/L5b-runs (runs)> datalad rerun fde26554f
[INFO ] run commit fde2655; (plot rois, traces...)
[INFO ] Making sure inputs are available (this may take some time)
unlock(ok): 05_visualization/240227J_Sim-Cre/displots/cell_01.png (file)
unlock(ok): 05_visualization/240227J_Sim-Cre/displots/cell_02.png (file)
unlock(ok): 05_visualization/240227J_Sim-Cre/displots/cell_03.png (file)
unlock(ok): 05_visualization/240227J_Sim-Cre/displots/cell_04.png (file)
unlock(ok): 05_visualization/240227J_Sim-Cre/displots/cell_06.png (file)
unlock(ok): 05_visualization/240227J_Sim-Cre/displots/cell_07.png (file)
unlock(ok): 05_visualization/240227J_Sim-Cre/displots/cell_08.png (file)
unlock(ok): 05_visualization/240227J_Sim-Cre/displots/cell_somata.png (file)
unlock(ok): 05_visualization/240227J_Sim-Cre/displots/cell_tuft.png (file)
unlock(ok): 05_visualization/240227J_Sim-Cre/rois/all_cells.png (file)
[1 similar message has been suppressed; disable with datalad.ui.suppress-simil[INFO ] == Command start (output follows) =====
Traceback (most recent call last):
File "/mnt/Data/L5b-runs/code/src/process2p/generate_plots.py", line 9, in <module>
from process2p.utils import *
ModuleNotFoundError: No module named 'process2p'
[INFO ] == Command exit (modification check follows) =====
[18 similar messages have been suppressed; disable with datalad.ui.suppress-similar-results=off]
run(error): /mnt/Data/L5b-runs (dataset) [python code/src/process2p/generate_plots...]
add(ok): 05_visualization/240227J_Sim-Cre/displots/cell_01.png (file)
add(ok): 05_visualization/240227J_Sim-Cre/displots/cell_02.png (file)
add(ok): 05_visualization/240227J_Sim-Cre/displots/cell_03.png (file)
add(ok): 05_visualization/240227J_Sim-Cre/displots/cell_04.png (file)
add(ok): 05_visualization/240227J_Sim-Cre/displots/cell_06.png (file)
add(ok): 05_visualization/240227J_Sim-Cre/displots/cell_07.png (file)
add(ok): 05_visualization/240227J_Sim-Cre/displots/cell_08.png (file)
add(ok): 05_visualization/240227J_Sim-Cre/displots/cell_somata.png (file)
add(ok): 05_visualization/240227J_Sim-Cre/displots/cell_tuft.png (file)
add(ok): 05_visualization/240227J_Sim-Cre/rois/all_cells.png (file)
[1 similar message has been suppressed; disable with datalad.ui.suppress-simil [18 similar messages have been suppressed; disable with datalad.ui.suppress-similar-results=off]
action summary:
add (ok: 28)
get (notneeded: 3)
run (error: 1)
save (notneeded: 5)
unlock (ok: 28)
What happened? - Apparently, I’ve lost the .venv directory during the hard reset which causes ModuleNotFoundError: No module named 'process2p'. määäh! This actually speaks for the use of a container. The problem with the container is that I have to rebuild it every time I update my code … annoying! (I learned that this statement might not be technically true, but since I’m only half-educated on this topic, I will leave it as it is for now.) I guess that’s the trade-off between efficiency and reproducibility. To illustrate this ‘highly complex’ dilemma with Deepseek’s smart-ass comment in a graph:
Fast Iteration ←──────────→ Reproducibility
↑ ↑
Convenience Containerized Certainty
The attentive reader might now also want to blame my decision to rewrite the history instead of creating a new commit which would have avoided the git reset operation and the loss of the uv environment. I guess also here there is a trade-off between maintaining a clean history and keeping development momentum:
Clean History ←──────────→ Development Flow
↑ ↑
Atomic Commits Rapid Iteration
For now I will just repeat the uv sync operation from above and add this code snippet to all my future scripts:
# Always ensure .venv exists
if [ ! -f "code/.venv/bin/python" ]; then
echo "WARNING: .venv missing! Recreating..."
cd code
uv sync
source .venv/bin/activate.fish
cd ..
fi
Run, Datalad, Run!
Meanwhile, the script has finished and left me with a set of successful [DATALAD RUNCMD] and an ocean of plots that I can swim in for the next few days :D Let’s push the results back to the main worktree! In my main worktree, I simply run git merge runs, and bahm, all the plots are there!
# back in the main worktree
/M/d/L5b (master)> git merge runs
Updating fde26554f..7f9f524f1
Fast-forward
05_visualization/240226I_Sim-Cre/Ketamine/displots/cell_01.png | 1 +
05_visualization/240226I_Sim-Cre/Ketamine/displots/cell_02.png | 1 +
05_visualization/240226I_Sim-Cre/Ketamine/displots/cell_03.png | 1 +
05_visualization/240226I_Sim-Cre/Ketamine/displots/cell_05.png | 1 +
05_visualization/240226I_Sim-Cre/Ketamine/displots/cell_06.png | 1 +
...
/M/d/L5b (master)> git log --oneline
7f9f524f1 (HEAD -> master, runs) [DATALAD RUNCMD] Plot rois, traces and distribution of F_corrected for mouse 240229K, Lisuride.
c2682b545 [DATALAD RUNCMD] Plot rois, traces and distribution of F_corrected for mouse 240229K, LSD.
923d34037 [DATALAD RUNCMD] Plot rois, traces and distribution of F_corrected for mouse 240229K, Ketamine.
80250575c [DATALAD RUNCMD] Plot rois, traces and distribution of F_corrected for mouse 240227J, Saline2.
...
I’m very happy! :D Also, meanwhile I did update my code repo with the code snippet above:
# still in the main worktree
/M/d/L/code> jj log
@ kxqpponr jiameng.wu@gmail.com 2025-12-04 21:18:05 fba5bff6
│ recreate venv inside the bash script # <- new commit
○ oqwzryko jiameng.wu@gmail.com 2025-12-04 20:18:57 git_head() 8af9e583
│ (empty) (no description set)
○ yknyuprl jiameng.wu@gmail.com 2025-12-04 20:18:57 runs 25eff5ce
│ update README.md
The change kxqpponr has been created in the main worktree during the plotting that happened in the runs worktree on top of the empty change from where I sent off the plotting jobs. Now I can simply clean up with jj abandon.
/M/d/L/code> jj abandon oq
Abandoned 1 commits:
oqwzryko 8af9e583 (empty) (no description set)
Rebased 1 descendant commits onto parents of abandoned commits
Working copy (@) now at: kxqpponr 7e551486 recreate venv inside the bash script
Parent commit (@-) : yknyuprl 25eff5ce runs | update README.md
/M/d/L/code> jj log
@ kxqpponr jiameng.wu@gmail.com 2025-12-04 21:21:45 7e551486
│ recreate venv inside the bash script
○ yknyuprl jiameng.wu@gmail.com 2025-12-04 20:18:57 runs git_head() 25eff5ce
│ update README.md
Beaauuutiful!
Conclude: All the magic in a nutshell
That was a very pleasant experience and quite a breakthrough in my personal data management journey. Between daily development chaos and production-ready reproducibility I found the sweetspot for a data analysis workflow that costs absolutely nothing in terms of time and disk space.
The beauty of this approach is its simplicity and efficiency:
- No data duplication: Worktrees share annexed files, so you don’t need to
datalad geteverything again - Parallel workflows: Develop in main worktree with Jujutsu or Git, process in clean worktree with DataLad
- Seamless integration: Merge results back with a simple
git merge - Low overhead: Environment setup happens once, then persists for all runs (unless you are enthusiastic time-traveler like me :P)
This isn’t a replacement for full reproducibility frameworks - for final publication runs, I still reach for containers and fully specified environments, and adapt something like the fairly big processing workflow. But for the 90% of my work that happens between initial exploration and final publication, this workflow hits the perfect balance.
Here is the destilled recipe for the workflow:
- Create a nested git worktree for your nested dataset on a new branch, here
runs - Set up the computational environment for this worktree
- Craft and test the
datalad runcommand invocation - Write a script that executes the datalad run command over multiple instances, here over subjects and experiments
- Make the update available on the branch/worktree
runs - Run the script in the worktree
runs - Merge the results into the main worktree
- Repeat 3-7 for the next iteration
Happy analyzing! 🧠